One of my favourite family board games is “Scotland Yard, the Hunt for Mr. X“. In this game you have 24 turns for a team of players to find the Spy (Mr.X) as he moves around the streets of London. I mention this because not only is this a great game, it’s also a rather clumsy, but accurate, way to introduce this blog topic, the X-chromosome.
I’ve been inspired to do this by gedmatch, the vendor-neutral website for comparing your DNA. Normally I’m not a big user of gedmatch, however I was surprised when I checked my X-DNA matches and saw that my top match was an unknown relative with a whopping 47.7cM matching segment on the X chromosome. To put this in context, the whole of the X chromosome is only 195.93 cM long AND my largest autosomal DNA match is only 42.4 cM over the other 22 chromosomes. This match was clearly worth investigating.
X-chromosome for Dummies
Before I go further on my own X-chromosome I think it’s worth giving a brief recap on the X chromosome (hereafter referred to by it’s more typist-friendly name of X). The X is part of the sex-chromosome. It’s what defines you as biologically either male or female. A male has only one, which he receives from his mother, whilst getting the rather puny Y from his father. A female has two Xs, one from their mother and one from their father. There is one other important difference here. The X that a mother gives to her children is recombined during meiosis. I’m afraid I’m rubbish at biology, if you are interested in the topic I’d highly recommend you do some background reading on meiosis and recombination.
In the mean time let me try and give you an executive summary. The X that a mother gives to her children is a mix of the two X chromosomes she has, this is what is referred to as “Recombination“. This mean that on average that X should be a mix of 50% of the X she received from her father and 50% of the X she received from her mother BUT it can be, and sometimes is, that she passes the whole of one of her X chromosomes to a child.
As you go back through the generations this process means two important things for a genealogist:
- You inherit your X or Xs from a specific subset of your ancestors. Trivia fact: The number of ancestors who a male inherits X chromosome DNA from in each generation follows the Fibonacci Sequence (1,1,2,3,5,8 etc. – assuming that the first 1 is the male’s own X…)
- In theory it is likely that you inherit your X in different proportions from different ancestors.
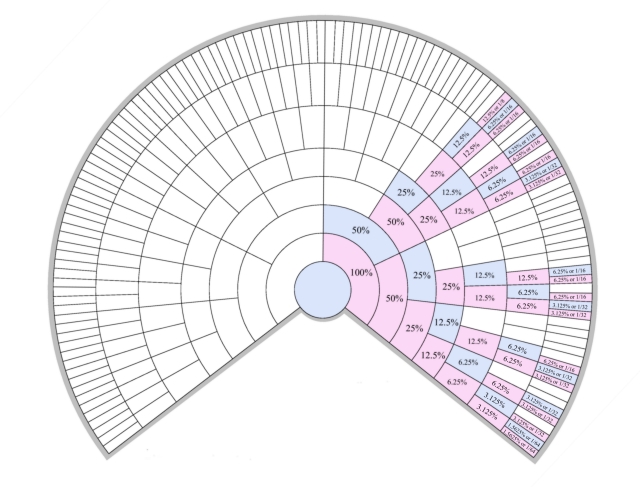
This process makes the whole understanding of X inheritance rather complex. Fortunately there are some nice charts for genealogists to use which help you understand this inheritance. I’ve taken the chart below from this website: www.thegeneticgenealogist.com/2009/01/12/more-x-chromosome-charts. There is obviously a slightly different chart for females, but I’m keeping that out of this blog, since the theory is challenging enough.
If I apply this to my family tree, then you can see who could have contributed to my single X chromosome, and what proportion the should have contributed.
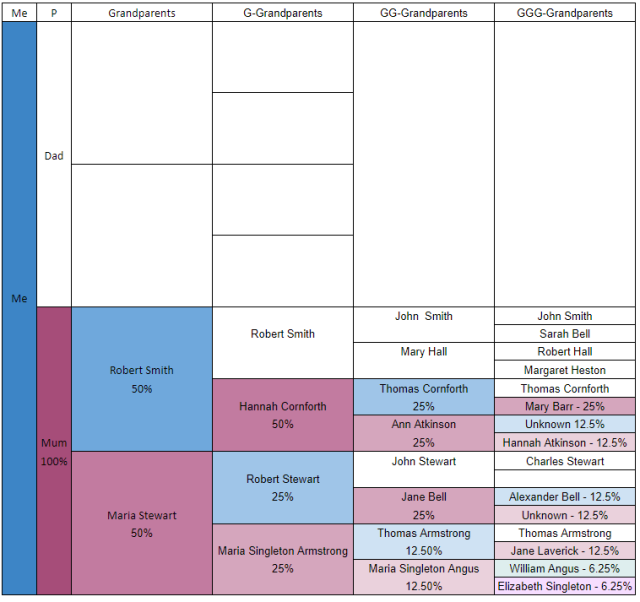
Hopefully this is clear, I’ve tried to shade the ancestors so it shows how much/how likely they contributed to my X.
As you can see, if you go back 5 generations there are only 8 people who could have contributed to my single X. Theoretically the largest contribution comes from Mary Barr (25%), whilst the lowest is from William Angus and Elizabeth Singleton who should have contributed only 6.25%. Contrast this with my regular autosomal DNA (the 22 pairs of non-sex DNA chromosomes) when all 32 ancestors should have contributed 3.125%.
There is a fly in the ointment. X chromosomes sometimes don’t seem to re-combine at all. It’s worth reading this article on the That Unruly X by genetic genealogist Roberta Estes. Having read this article I was curious to find out more about my own X.
Finding my X
The first step on my search was to compare my X against my brother. All four major DNA testers include SNP testing of the X as part of their autosomal testing. The exact number of SNPs they currently test (according to the ISOGG Wiki) are: AncestryDNA (28,892), FamilyTreeDNA (19,487) , 23andMe (18,091), and MyHeritage (17,889). Whilst the numbers differ I do not think there is a significant difference between the results the test companies provide. Since FamilyTreeDNA.com is where my brother and myself tested, and their website offers a chromosome browser I looked at their results first. Below is the result I got from comparing myself against my brother:

X-Chromosome comparison via FamilyTreeDNA. Orange segments are where we match
Remember here that my brother and myself have only on X each. So logically, every time our Xs make a switch from match to non-matching (and vice-versa) it is the result of a recombination event in one of our Xs. Without further information we don’t know if this was as a result of my X recombining, or my brother’s. If I look at this result it would appear that there are 9 such changes of state, implying 9 recombination events. To confirm this I also checked our results on gedmatch, which has a specific X one-to-one match-checking function. The result there was interesting. There were only 7 recombination events.

To confirm this I then went back to the familytreeDNA website and compared the two kits I have of my own DNA against each other. They should match over the complete length of the X. But the don’t, clearly there is a segment of the X that does not match:

X Chromosome comparison Myself match: Myself (orange) and my brother (light blue)
I can only assume that this is a SNP-poor part of the X that the FamilyTreeDNA matching mechanism cannot handle well. But at least it’s now clear, we are down to seven X recombination events, as the table below (taken from gedmatch) shows:
| Chr | Start Location | End Location | Centimorgans (cM) | SNPs |
| X | 12,192,147 | 33,957,251 | 37.0 | 3,244 |
| X | 45,064,135 | 114,485,507 | 47.3 | 5,216 |
| X | 125,242,332 | 141,847,221 | 24.2 | 1,979 |
| X | 147,291,278 | 154,545,424 | 16.9 | 1,057 |
Identifying Recombination events through Visual Phasing
The fun thing is we can now use the incredibly long segment of matching X from my x-match with an unknown shared ancestor to provide us with more information. This process is the same as used in “Visual Phasing“. As genetic genealogist Blaine Bettinger describes it:
“Visual Phasing is a process by which the DNA of three siblings is assigned to each of their four grandparents using identified recombination points, without requiring the testing of either the parents or grandparents”
I must admit I’ve tried to avoid Visual Phasing as a technique because it’s complex AND I don’t have the required number of siblings, or even first cousins that would help here, but in this case the long X match relation will work. Below are three one-to-one X comparisons done via gedmatch. The first “barcode” diagram is my brother and myself.
The second barcode is the X match between myself and my unknown relation. I’ve included the segment positions to make it clearer.
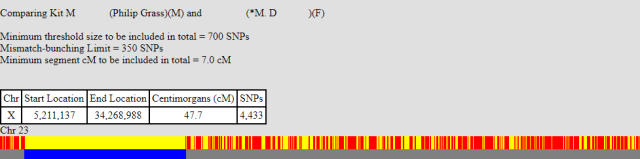
The third comparison: my brother compared to our unknown relation.
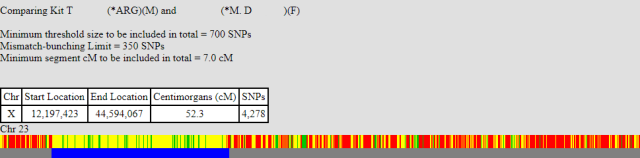
These three graphs show two important points:
- Looking from the start of the X chromosome; I match the unknown relation before and after the point where I first match my brother (position 12,192,147). This means that my X is very unlikely to have recombined at that point. My brother’s did.
- I stop matching the unknown relation at approximately the same position as I stop matching my brother (position 33,957,251 ). My brother continues to match the unknown relative. This means my X recombined at that point.
As a result I can now identify two segments each for myself and my brother:
| Who | Start Location | End Location | Centimorgans (cM) | SNPs |
| Me | 0 | 33,957,251 | ||
| Me | 33,957,251 | 45,064,135 | ||
| Brother | 0 | 12,192,147 | ||
| Brother | 12,192,147 | 45,064,135 |
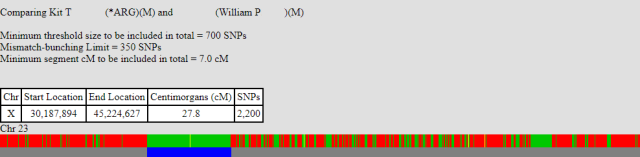
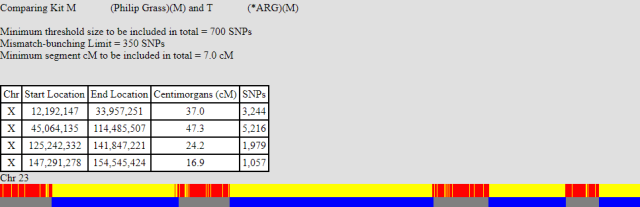
As further support for this, I’ve a couple more X comparisons to show. Again this is myself (10.6 cM), then my brother (27.8 cM), compared to a third party, namely WilliamP. Both our shared segments start at the same position (30,187,894). This does not fit with a our recombination event locations, so it must be that WilliamP’s X recombined at that point. You can see that my, shorter, shared segment finished at the position 33,957,251, which is the exact point I am expecting my X to have recombined.
The second point of note here is that my brother’s match with WilliamP stops around the 45,000,000 mark. This is approximately the same position as myself and my brother stop matching. This suggests, but doesn’t prove, that my brother had a recombination event at this point in the X.
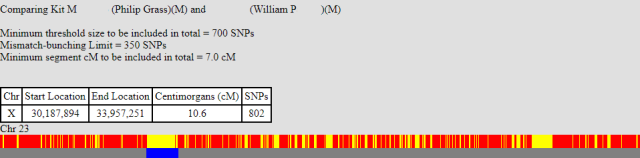
Completing the X with unknown relations.
Now that I’ve got my first two recombination events decided I want to try and see if I can completely map X. Unfortunately there are no other really large X-matches that I can use to determine recombination points. The good news is that some of the smaller c.20cM matches can help.
This is the first one, labelled NB. Below are the two barcode chart, myself and my brother, both compared to NB:
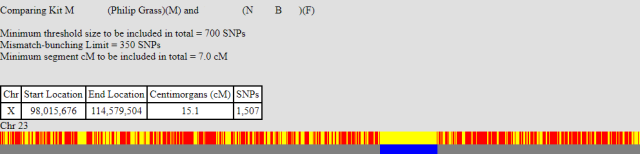
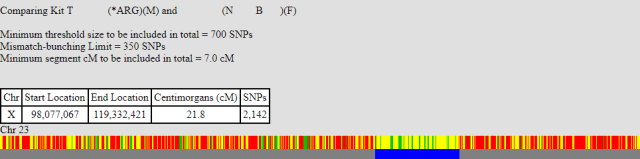
As you can see both start at approximately the same position around the 98,000,000 location. Whilst my brother’s match continues further, mine stops at 114,579,504, which is roughly the same position as myself and my brother stop matching (114,485,507). This means I must have had a recombination event at this point.
The next X-match that we can look at is one that for my brother hardly exists, whereas I have a 23 cM match. Note: I had to force gedmatch to use lower parameters to solve this part of the X. The matches, with an individual CG, are shown below:
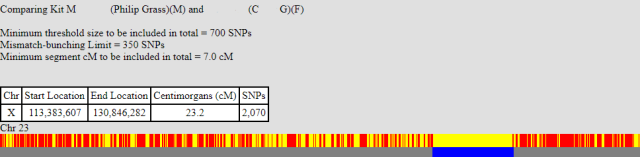
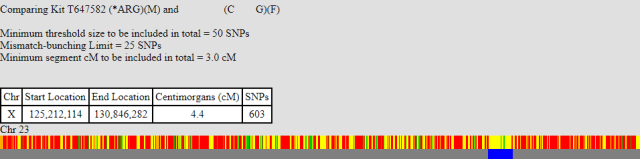
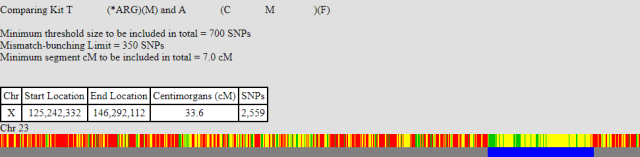
My brother only begins to match CG at the point 125,212,114. This is roughly the same position as my brother and I begin to share the same X. In other words he has a recombination event at this point.
Putting this information together we can now identify the following X lengths.
| Who | Start Location | End Location | Centimorgans (cM) | SNPs |
| Me | 45,064,135 | 114,485,507 | 47.3 | 5,216 |
| Me | 114,485,507 | 141,847,221 | ||
| Brother | 45,064,135 | 125,242,332 | ||
| Brother | 125,242,332 | 141,847,221 | 24.2 | 1,979 |
Update Sept 27th 2017 – A few weeks after first publishing this a second cousin very-generously uploaded her data to gedmatch. As a result I’ve re-written this last part of the article based on the additional information it provided.
Adding a known relative
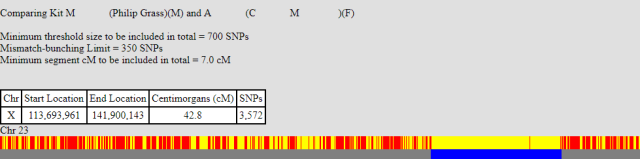
As per the update above I was lucky that a second cousin (refered to here as CM) uploaded her Ancestry data to gedmatch. Whilst we only share 42.8 cM the results are a real step forward for my X mapping. Below are her matches with myself and my brother.
These results enable me to do two things. Firstly I can identify the owner of another recombination point. This is around position 141,847,221 where I stop matching CM, whilst my brother continues, therefore my DNA recombined at that point. These results also support two of the previously-identified recombination events i.e. My brother had a recombination at position 125,242,332 and that I had one at 114,485,507.
Summing this up I have now the following identified Recombination points.
| Event Location | Who |
| 12,192,147 | Brother |
| 33,957,251 | Me |
| 45,064,135 | Brother (probably) |
| 114,485,507 | Me |
| 125,242,332 | Brother |
| 141,847,221 | Me |
| 147,291,278 | Unknown |
I’d love to be able to say that I found other X matches that allowed me to complete this exercise, however having combed through around 50 top X matches for both myself and my brother there are no other matches that clarify the last recombination event (position 147,291,278). After the top 50 matches the matching X lengths are around 10cM or less and so far, none cross the missing recombination points.
Putting it all together
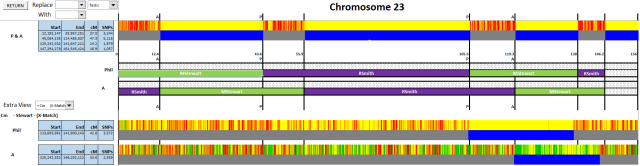
I’ve now discovered whose X DNA recombined at six points on the X chromosome and that rather elegantly both myself and my Brother have 3 each. More importantly, because I have a known second cousin who matches on the X, I can identify which grandparent provided the xDNA for virtually all of the chromosome. This is shown below.
Having identified which of my Grandparents provided which segments of DNA I can now use that information to narrow down my shared ancestor with any other match. Before this point I suspected that my match to MD (my mystery long match) was through the Bell family in Ulster sometime in the eighteenth century. This was based on our paper-trail research. This match fitted both our X inheritance patterns and is one of the more likely sources if you look at theoretical percentages of X inheritance. The good news is that my Bell ancestors are also ancestors of my Stewart grandparent and thereby supporting our paper-trail research.
Final Thoughts
In the past I’ve tended to ignore the whole idea of mapping your DNA segments to particular ancestors. both of my parents are dead and, since they were both a single-child I have no Uncles, Aunts or first Cousins to help identify DNA segments. I hope this blog post shows that even without much close family you can still identify the origins of at least the X part of your DNA. Just as important is the fact that the odd inheritance pattern of your X chromosome inheritance is actually helping you here.
If you are interested in the whole process of Visual Phasing then I would recommend you join this group on Facebook: www.facebook.com/groups/visualphasing/
No discussion of Visual Phasing is complete if it doesn’t mention the amazing spreadsheet that Excel-developer Steven Fox put together. If you are interested in Visual Phasing then you should try his spreadsheet. You can get hold of this from the Visual Phasing Working Group on Facebook. The magic of this spreadsheet is that it allows you to use Excel columns to align with recombination events from the gedmatch barcode diagrams. Below is a sample of the spreadsheet, showing the matches between my known Stewart relative and my brother/myself.
Finally, you may be wondering how often to expect the X chromosome to recombine. You can do no better than read this article by genetic genealogist Blaine Bettinger. This graph, taken from the articles, shows the distribution for the expected and actual number of crossover points in a study of 250 maternal-grandparents/grandchild family groups. My brother and myself have either 3 or 4 X DNA crossover events, which puts us towards the top end of the range, but not abnormally so.
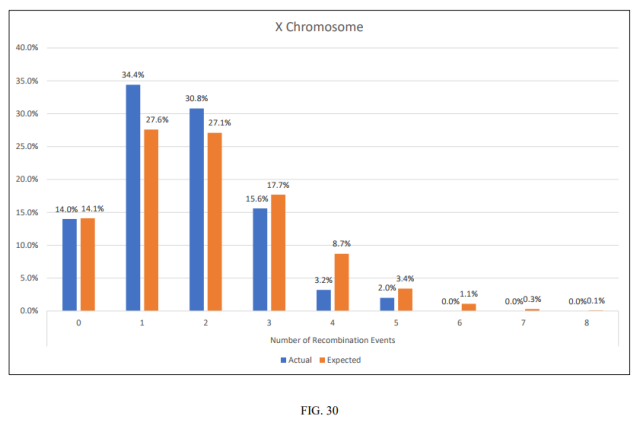
Expected and Actual X chromosome crossover rates – Taken from https://thegeneticgenealogist.com/wp-content/uploads/2017/02/Recombination_Preprint.pdf


Pingback: The moral maze of DNA testing | learnalittleeveryday
Hello Philip!
I enjoyed reading your post very much, thanks a lot!
There is just one little thing: some readers might get confused by the megabase numbers in the spreadsheet – they are slightly different than those at GEDmatch. And since you are using so many examples from GEDmatch, it would be better to use the GEDmatch numbers instead of the ones that the spreadsheet automatically assigns (you can change them manually).
According to the GEDmatch X-DNA comparison of you and your brother the crossovers are roughly at 12 – 34 – 45 – 114 – 125 – 142 – 147 Mb. However, in the spreadsheet image “X chromosome mapping by Grandparent” the megabase numbers for the crossovers are given as 12.4 – 43.6 – 55.9 – 105.5 – 119.3 – 138 – 146.2 Mb. Except for the first and the last crossover, the numbers are totally different (and it’s especially confusing when you look at the megabase numbers of the second and the third crossover).
Kind regards,
Eleonora
Hi Eleonora,
I’m glad you enjoyed the post – rather disappointingly it’s not one of the more popular posts.
Anyway, you’re right about the numbers -thanks for the feedback. I’ll edit them for consistency, although it might take a few days – it’s rather a long time since I was using the Visual Phasing tool, so I’ll need to re-learn it.